Peto's Paradox
 Whales, Elephants, and Bats
Whales, Elephants, and Bats
One of the many observations we have made is frequently referred to as Peto’s Paradox, so named after Richard Peto, who showed in 1975 that, per cell, mice had a higher rate of cancer than humans, and mused about the relationship between body-size and cancer rates. The Paradox, in a nutshell, comes from the simple fact that every cell in an animal’s body has the potential to become a tumor given the right set of mutations; furthermore, cells will steadily accumulate mutations throughout their lifetime. Thus, if you compare two species - large or small, long-lived or short-lived - you would expect the one with more cells, and a longer lifespan, to have an astronomically larger incidence of cancer. However, the fact that whales, elephants, and other long-lived and/or enormous animals have cancer rates comparable or below our own as humans, means that there must be some other mechanisms at play which compensate for the increased cancer risk; somehow, these species are able to either lower their own per-cell cancer rates, or else detect and destroy tumors near their onset.
This project was completed during my graduate career at the University of Chicago, and was pursued under the tutlage of Vincent J. Lynch, who is now at the University of Buffalo. My thesis, titled “The Role of Gene Duplication in Mediating Peto’s Paradox in Afrotheria and Chiroptera,” explores this question in great depth using a functional genomics and comparative biology approach that highlights the diversity of skills I developed in the Lynch Lab. What follows is the abstract to my thesis.
Cancer is a disease intrinsic to multicellularity. Within a species, body size and cancer risk are strongly correlated with cancer risk; between species, however, this correlation no longer holds. This phenomena, known as Peto’s Paradox, requires that species evolve cancer suppression mechanisms alongside increases in size and lifespan. Previous candidate gene studies looking at tumor suppressor duplications in large, long-lived species point towards a greater role of gene duplication in resolving Peto’s Paradox. Thus, in this thesis, I characterized gene duplications genome-wide in publicly-available genomes to determine if tumor suppressor genes in particular were enriched among duplicated genes in large and/or long-lived species. Then, to explore the functional consequences of these duplicates, I selected two hits in large, long-lived species - LIF in the African Savanna Elephant (Loxodonta africana) and TP53 in the Little Brown Bat (Myotis lucifugus) to characterize in primary fibroblasts, and determine their effects on cell cycle and cell death in response to stress.
Expanding upon previous candidate-gene-based surveys of gene duplication in Elephants, we used a Reciprocal Best-Hit BLAT strategy to obtain copy numbers of all protein-coding genes in Atlantogenatan genomes to see if there is any correlation between the copy number of duplicates and changes in body size along the phylogenetic tree. From an initial set of 18,011 protein-coding genes in hg38, we identified a median of 13,880 genes in Atlantogenatan genomes, of which a median of 940 genes are duplicated. We find that, just as body size fluctuates throughout Atlantogenata, genes involved in tumor suppressor pathways are also duplicated throughout the phylogenetic tree. Extant species of elephants, however, show active transcription of both canonical and duplicated copies of tummor suppressors that duplicated prior to and during their sudden increase in body size, suggesting that the duplication of tumor suppressor genes facilitates the evolution of increased body size by compensating for the increased cancer risk.
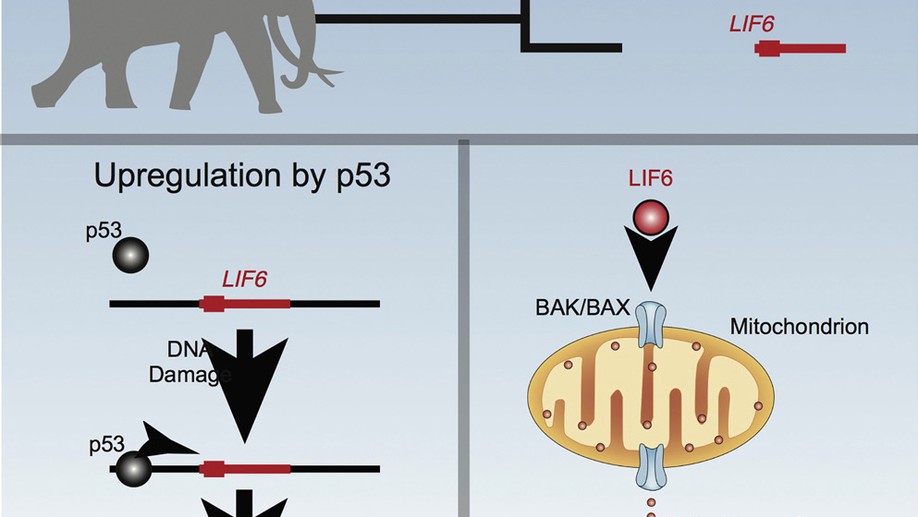
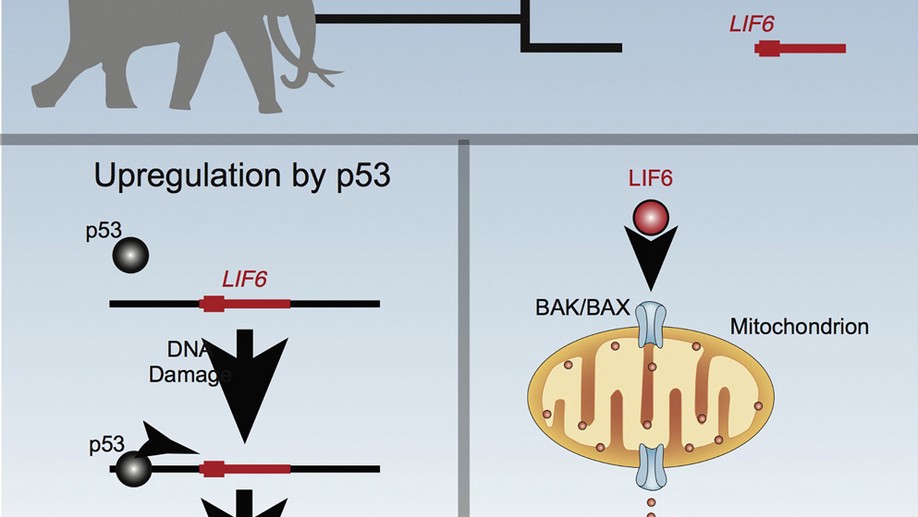
The resurrection and re-functionalization of a leukemia inhibitory factor pseudogene (LIF6) with pro-apoptotic functions in elephants and their extinct relatives (Proboscideans) may have played a role in resolving Peto’s Paradox. LIF6 is transcriptionally up-regulated by TP53 in response to DNA damage, and translocates to the mitochondria where it induces apoptosis. Phylogenetic analyses of living and extinct Proboscidean LIF6 genes indicates its TP53 response element evolved coincident with the evolution of large body sizes in the Proboscidean stem-lineage. These results suggest that re-functionalizing of a pro-apoptotic LIF pseudogene may have been permissive (though not sufficient) for the evolution of large body sizes in Proboscideans.
In the long-lived bat, Myotis lucifugus, I describe the only known full-locus duplication of TP53 in nature, which may play a role in shaping its unique stress response. This bat has 8 copies of TP53, a central regulator of the DNA damage response present in all living organisms. A syntenic duplication of TP53-WRAP53 has occurred within its genome, leading to two copies of TP53 that have conserved both regulatory and transcriptional functionality. Relative to 4 other closely related bat species (M. evotis, M. thysanodes, M. yumanensis, and E. fuscus), the Little brown bat demonstrates a unique resistance to both DNA damage as well as generalized oxidative stress, which matches the phenotypic changes induced by a transgenic TP53 locus duplication in a previously-described mouse model.
Overall, these results suggest that gene duplication plays an important role in Peto’s Paradox; while individual cases of tumor suppression duplication can facilitate the evolution of increased lifespans and body sizes, the suppression of cancer risk likely requires a concert of genetic changes. This fits in with a polygenic or omnigenic model of cancer resistance and tumorigenesis, and suggests that large-scale integrated functional genomic approaches to studying Peto’s Paradox may provide greater clarity into how large, long-lived species suppress their increased cancer risk.